Contents
- Index
- Previous
- Next
Setting up FindSites
This optional step in setting up the analysis will permit you to assess informative sites for evidence of recombination. For a description of this method, see:
Robertson,D.L., Hahn,B.H., and Sharp,P.M. Recombination in AIDS viruses. J Mol Evol 1995;40(3):249-59
The first step is to select 4 sequences, one to represent the Query (putative recombinant), 2 References (potential parentals of the recombinant), and a fourth "Other" sequence to function as a control.
Sequences are selected by using the Combo boxes on the SeqPage panel:
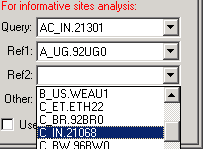
Groups versus Individual sequences:
Because SimPlot can generate consensus sequences from groups, you also have the option of using groups rather than individual sequences for informative sites analysis. This has the advantage of reducing the effects of variation (artefactual or biological) in individual sequences; however, [as with all such analyses] if groups are assembled carelessly they will yield erroneous results.
There is a checkbox at the lower-right corner to allow you indicate your preference:
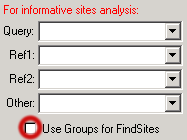
Having made that selection, the Combo boxes now display only the groups:
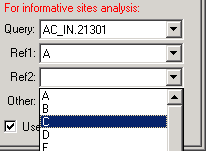
Note that when using groups in informative sites analysis, there is an option to select the degree of conservation required to include a site in the analysis. Setting this to 100% will require that a site is homogeneous in each of the 4 groups. A 50% setting will result in use of a simple consensus for each group.
Once you have made selections in ALL FOUR Combo boxes, you will be able to assess informative site partitions in subsequent steps (e.g. see the next page in this tutorial).


