Contents
- Index
- Previous
- Next
Bootscanning
Bootscanning was first described by Mika Salminen:
Salminen MO, Carr JK, et al. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995 Nov;11(11):1423-5
The basic principle of bootscanning is that mosaicism is suggested when one observes high levels of phylogenetic relatedness between a query sequence and more than one reference sequence in different genomic regions. Using default values for our demonstration data, we obtain:
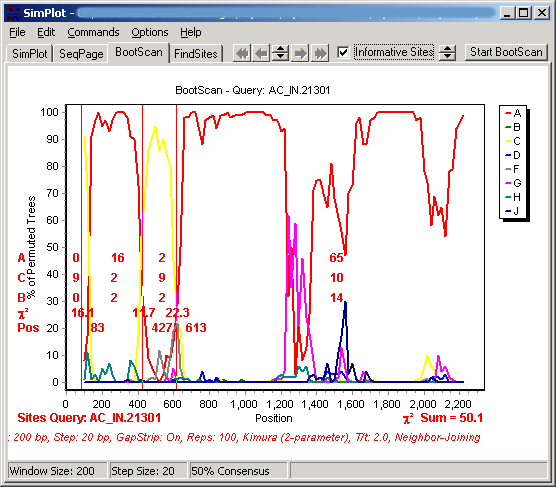
When the bootscan is completed, we are notified that a number of groups appear to be potentially mosaic. Clicking on each of these in the legend allows the user to view the bootscan plots for each of these as a query. The notification regarding possible evidence of recombination, and the threshold for this notification, is available in Preferences under the Options menu.
Note: The raw values of the bootscan can be saved in comma-delimited format from the File menu.
